SIM
The 3-dimensional structured illumination microscopy (3D-SIM) belongs to the group of super resolution microscopes some of which have been awarded with the Nobel Price 2014 for chemistry.
In order to get a high resolution image a distinct light pattern is laid over the focal plane of the sample and is repeatedly shifted and rotated while acquiring images. These images are consequently processed and aligned with a specialized algorithm. These acquisitions are done across all planes of the specimen. This allows a reconstruction of a 3D stack.
If one applies this technique to biological samples that are stained for specific proteins or structures, one can study localization, interactions and various other biological processes.
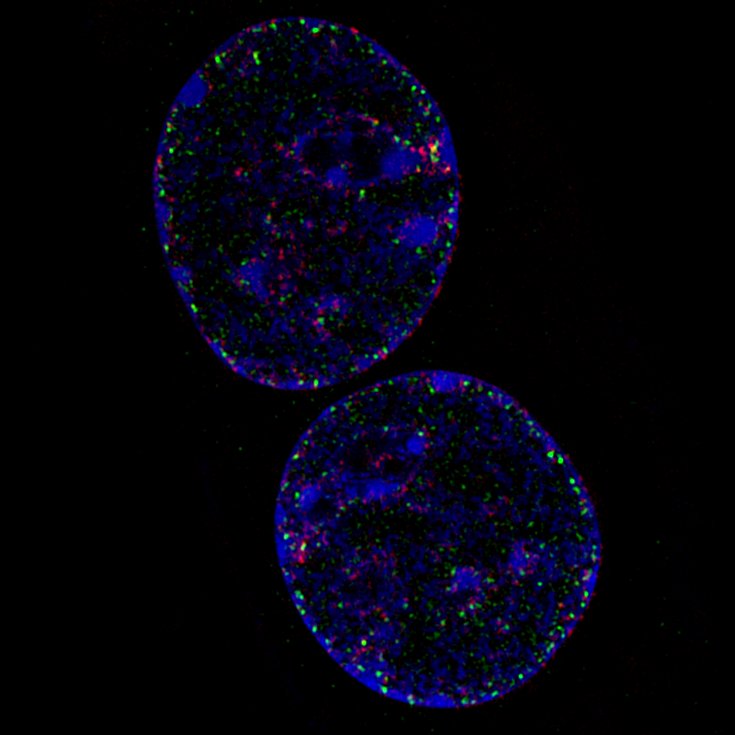
In the course an EdU-chasing experiment was performed which allows tracking of the DNA replication progress.
Video 1: 3D stack of a C2C12 cell in mid-S- Phase of the cell cycle|DNA (Blue), EdU (green), PCNA (red)| EdU are synthetic nucleotides which were incorporated 20 minutes prior to fixation, i.e. green staining marks the position of the replication fork 20 minutes ago. PCNA is a DNA-binding protein located at the replication fork, i.e. red staining marks the actual position of the replication fork at the time of fixation.
An additional experiment shows the exact position of the nuclear pore complexes within the nuclear envelope. These precise localization studies are facilitated only by high resolution microscopy techniques like 3D-SIM.
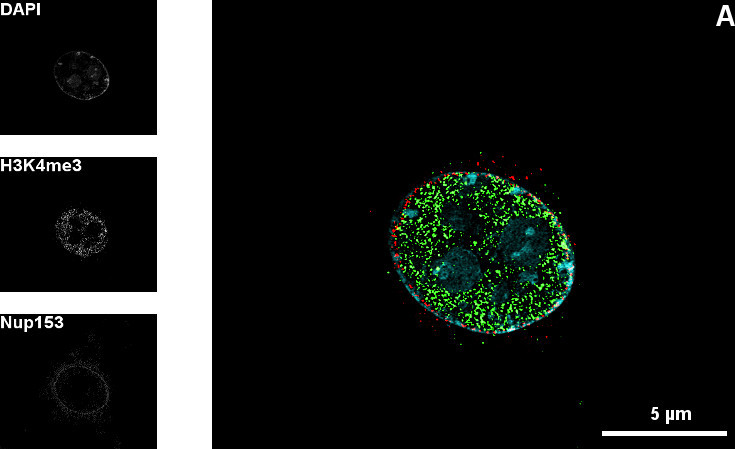
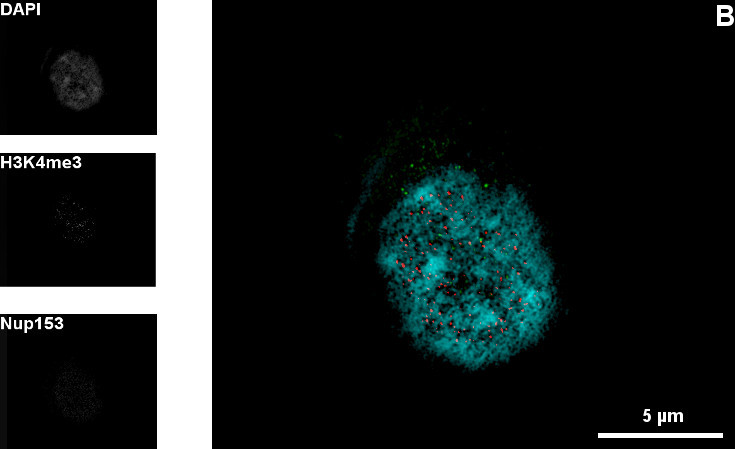
Figure 1: Mid-section (A) and top-section (B) of the nucleus of a C2C12 cell |DNA (Blue), H3K4me3 (green), Nup153 (red)| A and B show the nuclear pore complexes within the nuclear envelope in different optical sections through the nucleus. The pores allow different molecules to pass into the nucleus. The 3D-SIM technique shows the pores in a cellular substructure with a diameter of about 100 nm. The gaps in the DAPI counterstaining correspond with the position of the nuclear pore complexes. The sample is also stained against H3K4me3. This abbreviation stands for a specific histone modification.