FRAP
In order to analyze the mobility of different proteins inside of the nucleus, HeLa cells were transfected with various constructs (GFP, GFP-H2A, GFP-PCNA and GFP-Dnmt1). A small area of the nucleus was photobleached with a laser pulse and the recovery of the respective protein was detected via Spinning Disc Confocal Microscopy within a predefined timeframe. This way the mobility of the protein of interest as well as its strength of interaction with another structure (e.g. chromatin, membrane proteins) could be validated.
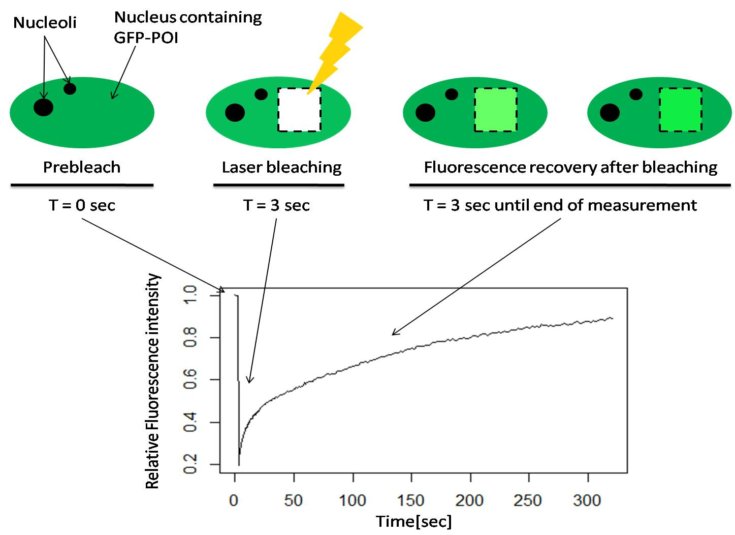
Fig. 1 Schematic illustration of Fluorescence Recovery After Photobleaching (FRAP) of a GFP tagged protein of interest.
Click here for video of a FRAP PCNA experiment
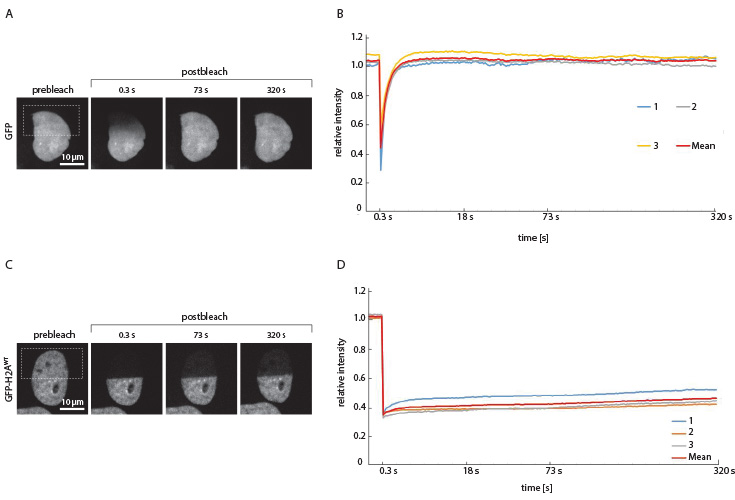
Fig. 2: FRAP of GFP in the HeLa cell nucleus. A + B: As GFP is not involved in cellular processes it freely diffuses through the cell and can be used as a positive control for FRAP experiments. After bleaching of the region of interest, fluorescence recovery was reached after approximately 4 seconds, revealed by quantitative analysis of the relative light intensity with respect to the time.
C + D: FRAP of GFP-H2A in the HeLa cell nucleus. In comparison to freely diffusing GFP, the GFP bound to the Histon H2A shows a much longer recovery time after bleaching. It could be concluded that H2A does not occur freely in the nucleus, but rather is bound to a static structure. This is in agreement with the observation, that H2A is part of the histon octamer with the DNA wrapped around, leading to relative immobility. Fluorescence could not be recovered.
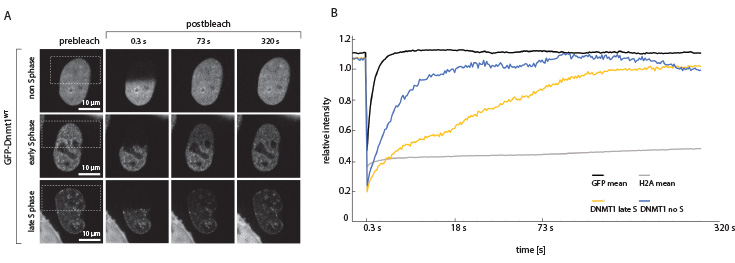
Fig. 3: FRAP of GFP-Dnmt1 in the HeLa cell nucleus. A + B: After replication the hemimethylated newly synthesized DNA is methylated by the maintenance DNA methyltransferase 1. During late S-phase Dnmt1 is located proximal to the replication fork. As expected the fluorescence recovery is delayed compared to the non S-phase cell, where Dnmt1 diffuses freely in the nucleus.
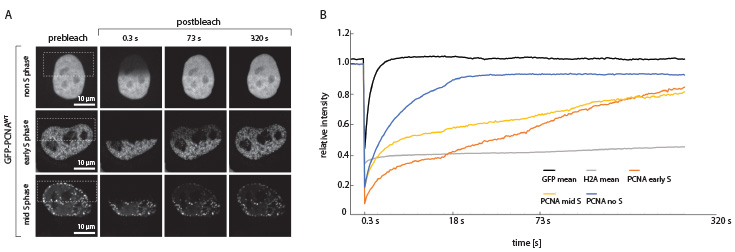
Fig. 4: FRAP with GFP-PCNA in the HeLa cell nucleus. PCNA is the sliding clamp that guides the DNA polymerases δ and ε along the DNA strand during replication. It is also extensively posttranslationally modified and thereby serves as a signaling platform regulating replication processivity. In S-phase cells PCNA is bound to the DNA polymerase at replication forks, whereas after replication it can diffuse freely. This is in agreement with the FRAP experiments, showing a slower fluorescence recovery for the S-phase cells compared to the non-S-phase cell.

