Confocal
The significant advantage of the scanning confocal microscopy (LSCM) in contrast to the standard widefield LSM comes with a pinhole. Traditional widefield epi-fluorescence microscope objectives focus a wide cone of illumination over a large volume of the specimen, which is uniformly and simultaneously illuminated. A majority of the fluorescence emission directed back towards the microscope is gathered by the objective (depending upon the numerical aperture) and projected into the eyepieces or detector. The result is a significant amount of signal due to emitted background light and autofluorescence originating from areas above and below the focal plane, which seriously reduces resolution and image contrast. LSCM focuses the laser illumination source through a lens to a very small point at the focal plane. The fluorescent emission is filtered through a small pinhole before reaching the detector, which minimizes florescent emission from out of focus fluorophores. The scanning is achieved by two mirrors driven by galvanometer motors. One of the mirrors moves the beam from left to right along the x lateral axis, while the other translates the beam in the y direction. After each single scan along the x axis, the beam is rapidly transported back to the starting point and shifted along the y axis to begin a new scan. Participants of the Bioimaging Course 2016 were able to experience this microscopy method first hand.
For the LSCM, two slides were prepared previously in the practical. Both slides were seeded with C2C12 cells (a mouse myoblast cell line). Therefore coverslips with a thickness of 0.170±0.005mm and with an area of 18x18mm were used. The immunostaining was different for each sample. Slide 1 was incubated with primary antibodies binding H3K4me3 (specific histone type, common in tightly packed chromatin) and Nup153 (a nucleus pore protein). Prior to the antibody application slide 2 was incubated with EdU (5-Ethynyl-2´-deoxyuridine, a thymidine analogue) which afterwards is detected with a fluorescent azide. Additionally both samples were counterstained with DAPI.
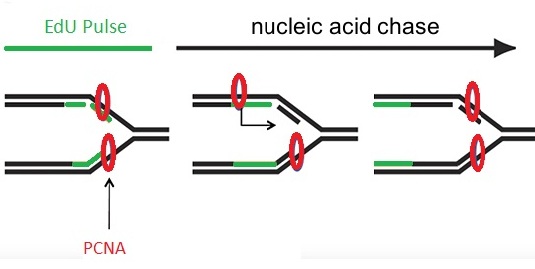
Figure 1: Schema of EdU pulse-chase experiment.
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a scaffold to recruit proteins assisting DNA replication, DNA repair, and chromatin remodeling. The region of DNA labeled with EdU (green, labeled with 6-FAM-Azide) starts at the replication fork and grows as the fork progresses along the DNA helix. Newly synthesized DNA is associated with replication proteins PCNA (red, labeled with rat anti PCNA). Incubation with a brief pulse of EdU followed by a chase of nucleic acid leads to EdU incorporation in a small segment of DNA that becomes more distant from the moving replication fork as the chase time increases.
Images were taken by each group for their self made samples guided by an instructor. After a short introduction about LSCM first x,y axis images were taken and then a z series was recorded. The participants learned about all possibilities to optimize the microscope settings for their certain specimen. As they spotted a suitable cell, the next step was to center and focus the selected cell by using the eyepiece. To configure the exposure time, photomultiplier sensitivity and laser power for each channel a specific software was used. Images were taken with three different channels for DAPI, H3K4me3, PCNA, EdU and Nup153. Coupled to a secondary antibody Alexa594 binds to Nup153 and PCNA, so does Alexa488 to H3K4me3 and EdU. For a higher photon count the sampling quantity of each point can be increased.

Figure 2: Laser scanning confocal microscopy from Leica SP5
Immunostaining of nucleoporin 153 (Nup153) and histone 3 lysin 4 trimethylation (H3K4me3) in a C2C12 cell (mid section)
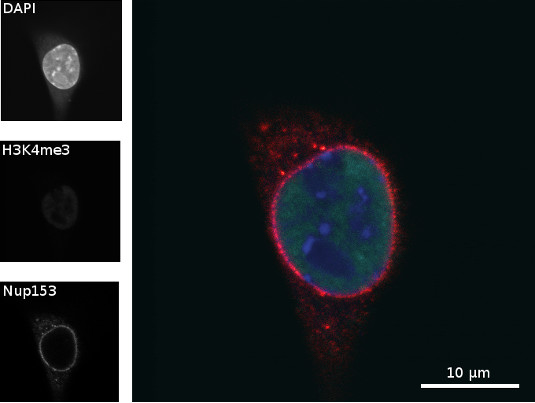
Figure 3: Chromatin of a C2C12 cell was stained with DAPI (blue), H3K4me3 (green) was immunostained with rabbit anti H3K4me3 (primary antibody) and goat anti rabbit Alexa488 (secondary antibody), and Nup153 (red) was immunostained with mouse anti Nup153 (primary antibody) and donkey anti mouse Alexa594 (secondary antibody). Scale bar = 10 µm
Figure 3 shows a middle section of C2C12 cells, in which nucleoporin 153 is located at the nucleus membrane, where the nuclear pore complexes are supposed to be. In the nucleus euchromatin and heterochromatin are located. In heterochromatin the DNA is more tightly packed than in euchromatin and thus is brighter when stained with DAPI, because there is more DNA. The image shows that the trimethylation at lysine 4 of histone 3 (H3K4me3) co-localizes with euchromatin, since in heterochromatin the DNA is so tightly packed, less histone methylation can be found.
Immunostaining of nucleoporin 153 (Nup153) and histone 3 lysin 4 trimethylation (H3K4me3) in a C2C12 cell (top section)
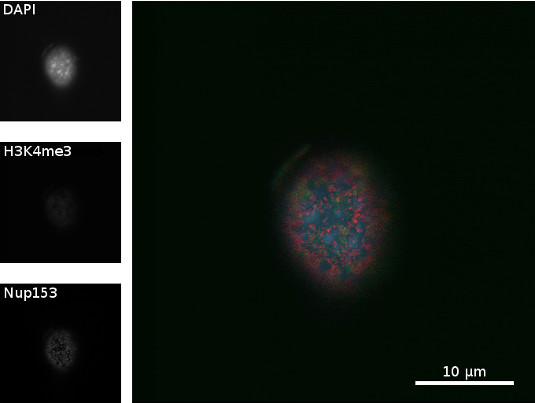
Figure 4: Chromatin of a C2C12 cell was stained with DAPI (blue), H3K4me3 (green) was immunostained with rabbit anti H3K4me3 (primary antibody) and goat anti rabbit Alexa488 (secondary antibody), and Nup153 (red) was immunostained with mouse anti Nup153 (primary antibody) and donkey anti mouse Alexa594 (secondary antibody). Scale bar = 10 µm
Figure 4 shows a superior view of C2C12 a cell, where Nup153 spread over the nucleus. Since it is the section of the top of the cell, the nuclear pore complexes can be seen on the membrane. Histone and chromatin show weaker signal because they are underneath the membrane.
Immunostaining of EdU Click-iT pulse-chase experiment and PCNA in C2C12 cells
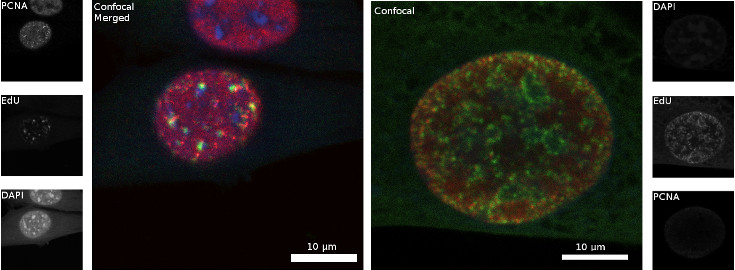
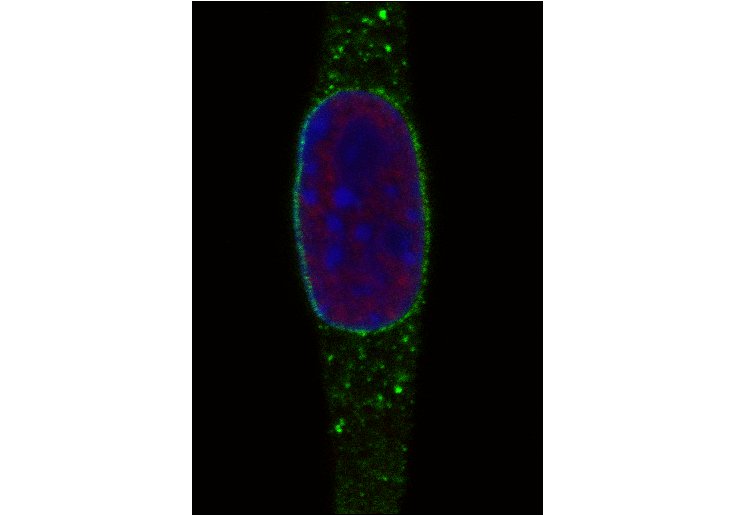
Figure 5: C2C12 cell nuclei where the chromatin was stained with DAPI (blue), EdU was stained with 6-FAM-Azid (green), and PCNA (red) was immunostained with rat anti PCNA (16D10) (primary antibody) and goat anti rat Alexa594 (secondary antibody). Scale bar = 10 µm, left: late S phase; right: early to mid S phase.
The left panel in figure 5 shows the result of EdU Click-iT pulse-chase experiment while the C2C12 cell was in late S phase, whereas right panel shows the incorporation of EdU during replication in early to mid phase. During the late S phase, constitutive heterochromatin, where the chromatin is most tightly packed, is replicated. Constitutive heterochromatin is mostly formed of gene poor and tandem repeat regions at pericentromeres. The cell at the left panel incorporated EdU during the replication of constitutive heterochromatin in late S phase, which can be seen in the few bright dots. Since the replication was continued without EdU, PCNA moved further on the replication fork and is distributed throughout the nucleus at the moment of fixation. In the right panel the cell was during early to mid S phase, where euchromatin and facultative heterochromatin are replicated, when EdU was incorporated. That is why bright but smaller green dots of 6-FAM-Azid stained EdU spread across the nucleus can be seen.
The data is evaluated with different software (e.g. Fiji, GIMP, Amira). With a z series it is possible to create 3D images by aligning several x,y-axis images. The participants were extensively instructed in the usage of those programs.